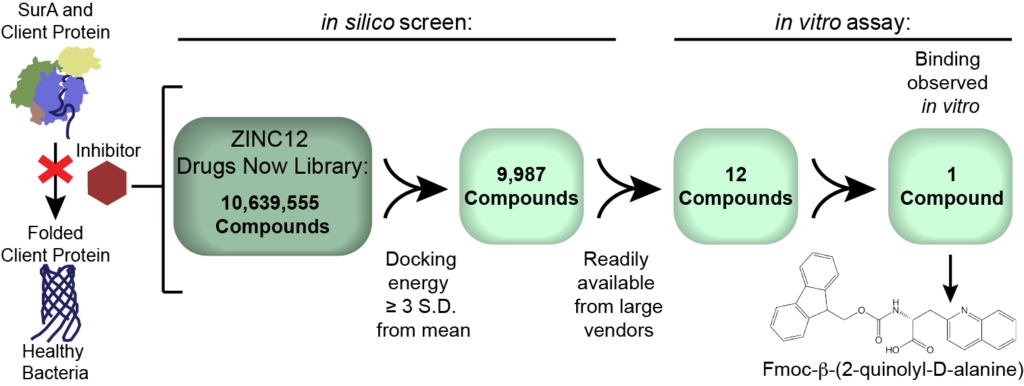
Our lab focuses on a chaperone protein, SurA, found in the periplasmic space of gram-negative bacteria. Chaperone proteins assist in folding other proteins into their functional states. SurA acts as a gatekeeper between the interior of a bacterium and the external environment by being responsible for the proper folding of secreted proteins and toxins as well as the overall maintenance of the cell membrane. Understanding the mechanism of SurA chaperoning will allow us to strategically inhibit the chaperone and reduce the viability of gram-negative bacteria, potentially making them more susceptible to already existing antibiotics.
We used computational methods to dock small molecule libraries to the crystal structure of SurA. These methods ultimately make a prediction about the binding affinity (how tightly a small molecule and SurA fit together), which can be used to predict whether or not a molecule is an effective inhibitor of the chaperone. We have completed a screen of the ZINC12 library (>10,000,000 compounds) and determined which compounds bind best by setting a threshold of binding affinity three standard deviations from the predicted mean binding affinity. We utilized Oberlin’s supercomputing cluster, SCIURus, to dock the library and ridge regression analysis to sort the hit compounds by common functional groups weighted by the compound’s binding affinity. We found the most stringent method of reducing the number of hit compounds was to identify those that were commercially available at high purity from trusted sources.
We tested a subset of these hit compounds using an in vitro competitive binding assay. Using purified SurA protein and fluorescently labeled peptides known to bind SurA, we have developed and optimized a fluorescence anisotropy-based assay.
In the fall of 2018, our lab published an article titled “Identification of inhibitors of the E. coli chaperone SurA using in silico and in vitro techniques” in Bioorganic and Medicinal Chemistry Letters highlighting these computational and biochemical efforts.